Who's who?
The library and the module have evolved over the years, raising some confusion between all the names used to label the various pieces. This page tries to clarify this situation.
"MED" can (unfortunately) refer to:
- MED file format: the file format used to save a mesh (".med" extension)
- MED-file library: the C++ library developped by EdF R&D (and provided with SALOME) to read/write MED file (warning: for advanced users only!)
- MEDCoupling: the (relatively) high level API to deal with mesh and fields in memory
- MEDLoader: Writing, reading MED files MEDLoader: part of the library dedicated to file I/O = a more user-friendly API than the MED-file library API
- Remapper: part of the library dedicated to interpolation/projection methods
- SALOME’s MED module (GUI point of view): a graphical client in the SALOME main application, providing a graphical interaction with part of the library ; for this please refer to the MED module documentation
- and finally ParaMEDMEM, for the projection operations and field transfers in parallel
The most common confusion is between the MED library (what you are reading at present) and the MED-file library ("MED fichier"). The MED-file library is part of the prerequisites of the MED libary, and its only purpose is to read and write MED files. This is a low level API written in C, and giving a fine-grain access to the structure of the MED files (.med). The architecture diagramm below details those points further.
Another source of common confusion is that all the standard (sequential) MEDCoupling/MEDLoader API lies in the ParaMEDMEM namespace. This is quite unfortunate but due to historical reasons. The true parallel functionalities of the MED library are detailed here: Parallelism, but are still often called the ParaMEDMEM part of the library.
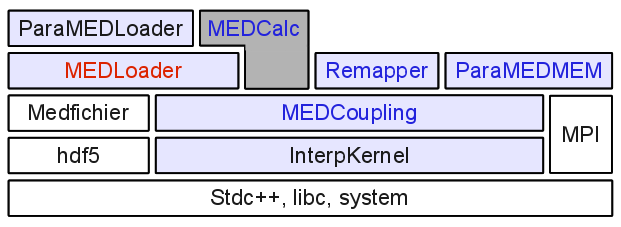
Architecture
The figure below represents the layer structure of the packages of the library.
- each element depends on the blocks it covers (fully or partially).
- White blocks represent system or external dependencies.
- the MEDCalc block, with gray background color, is the Graphical User Interface (please refer to the MED module documentation).
- red-colored text identifies code with Swig interfaces (API available in Python)
- blue-colored text identifies code with both Swig interfaces and CORBA layer.
The fundamental set (blue background) consists in three atomic libraries:
- MEDCoupling that describes data structures used for cross process exchange of meshes and fields.
- MEDLoader and ParaMEDLoader that provides I/O functions to the MED file format with sequential and parallel processing, respectively. Those are built on top of the MED-file library.
- interpolation tools that provides mathematical structures and algorithms for interpolation and localization. It is implemented in three blocks: InterpKernel, Remapper and ParaMEDMEM (Remapper with parallel processing).