Version: 8.3.0
MEDLoader is a package in charge of loading from a file or write to a file in MED format a MEDCoupling data structure. The fact that these functionalities are not merged in the MEDCoupling library (separated .so files) is explained by a willingness of reducing as much as possible the dependencies of this library.
As a MED file can combine several MEDCoupling aspects in one (for example meshes in MED file on more than one dimensions with families and groups) the API of MEDLoader is much more rich than simply read and write.
MEDCoupling mesh is not as rich as a MED file mesh, and a MEDCoupling field is not as rich as a MED file field. But it is possible to emulate with a very good fidelity a MED file mesh and a MED file field with a collection of MEDCoupling instances for each.
MEDLoader module offers two different approaches to perform Read/Write from/to MED file.
Whatever the approach(es) you choose, it is advisable to know main concepts of MED files that are quickly reminded here.
A specific page dedicated to the basic API is available here.
This approach is less close to MED file concepts, but closer to MEDCoupling concepts.
So, basic API is simpler, as shown by method MEDCoupling::WriteUMesh that needs no special knowledge about MED file concepts to interact with MED files.
This API is in the form of a list of public static functions directly in the namespace MEDCoupling.
This simplicity has a cost, the I/O are not (cannot be) optimized.
As MED file concepts are more complex than MEDCoupling concepts, this approach is not the most relevant for specific MED file objects read/write.
A specific page dedicated to the advanced API is available here.
This approach is the closest to MED file. By using this advanced API approach the user will manipulate classes that represent MED file concepts.
It implies that the user should be informed about the MED file concepts, that do not exist in MEDCoupling. For example :
This is typically the case for a user that wants to precisely set/get mesh/group/family groups set on different level.
This is why a set of classes corresponding to memory representation of MED file concepts is proposed by advanced API approach.
So All information contained in file is represented by advanced API class instances.
The level of coherency check is variable across methods, to let to the user the maximal capacity of modification of its MED file data in memory.
This API is particularly recommended :
The two approaches are NOT opposed, they are compatible each other so it is possible to mix them.
Typically it is possible to read rich information of a complex MED file using advanced API in read mode, postprocess this input then get a simpler MED file model, and write it to a simple output MED file using basic API.
Here we will describe some of the basic concepts of MED files in order to use the best methods proposed by MEDLoader API.
First of all MEDLoader will not read MED files whose version is strictly lower than 2.2.
For new comers in MED file world some of basics principles are recalled in the following graphic :
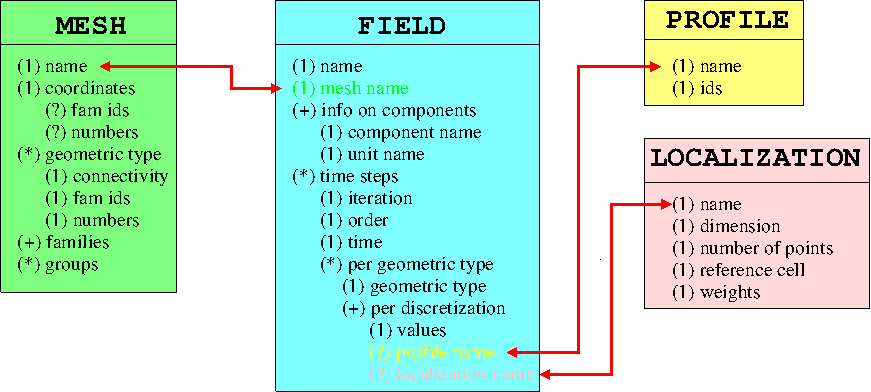
Inside the parenthesis, there is multiplicity :
Each box is independent in MED file format during read/write session.
Boxes instances are linked each other only by red arrows using string as discriminating key. It implies that empty names in basic concepts objects of MED file are forbidden.
There can be as many instance of boxes as wanted in a MED file.
As it can be seen in MED file world, fields and meshes are sorted by geometric type.
This specificity leads to a constraint during writing phase because some mesh operations may significantly modify the organization of geometric types during mesh/field operations.
Here are some of operations that can alter the geometric type order of cells:
As it has been seen above, all big arrays in fields and meshes (except coordinates) are sorted by geometric type, without any awareness of the dimension.
For example an unstructured mesh in MED file world can lie simultaneously on MED_TRI3, MED_POINT1, MED_POLYHED, MED_TETRA4..., which is impossible in MEDCoupling for manipulation reasons.
To connect the MED file world to the MEDLoader/MEDCoupling world the notion of relative mesh dimension has been introduced in MEDLoader.
This concept of relative mesh dimension is used frequently in the MEDLoader APIs (basic and advanced).
To explain the semantic of relative mesh dimension let's take the example of a mesh called myMesh in a MED file, containing MED_TRI3, MED_POINT1, MED_POLYHED, MED_TETRA4.
For each geometric type on which myMesh is defined the mesh dimensions are :
The mesh dimension of myMesh is equal to 3 ( max(2,0,3,3) ). The relative mesh dimension is equal to the difference between mesh dimension of geometric type and the mesh dimension of the whole MED file dimension. It leads to the following relative mesh dimension :
In MEDLoader all geometric information are then grouped relative dimension per relative dimension. It leads to the following geometric sorting of MED file data structure of myMesh :
The mesh dimension of myMesh is 3. The relative mesh dimensions available are 0, -1 and -3.
As it has been seen previously in for meshes, the values of fields are sorted by levels too.
The principle is the same than those explained for meshes. The only difference is in the fact that it is possible for fields on cell and fields on Gauss points that mesh dimension of underlying mesh of a field is not always (but very often) equal to the dimension of geometric types on which this field is defined.
So it is advised, to compare the non empty level of a field and of its underlying mesh before trying to request heavy data from a MED file.
As seen above, fields in MED file can be defined on different time steps.
But there is a major difference with MEDCoupling concept in time steps. MEDCoupling focuses on the float value of time for interpolation reason.
MEDLoader: Writing, reading MED files MEDLoader and MED file focus on pair of integers to precise a specific time steps.
This difference of point of view can be seen in the API where the order of returned parameters in python differs from MEDCouplingFieldDouble::getTime to MEDFileField1TS::getTime.
In MED file and so in MEDLoader the time steps are identified by a pair of integers called :
Order refers to sub iteration id, that is by default set to -1 in case of non use. A physical time with float type is attached to this pair of integer.